
 | Cluster Information |
| Prev | Using Klusters | Next |
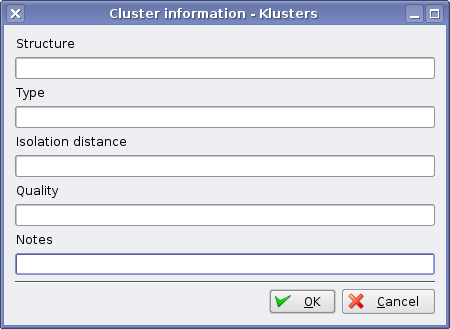
Klusters allows to specify important information about the clusters, namely:
Structure - the brain structure where the unit was recorded.
Type - the type of neuron, e.g. pyramidal cell.
I.D. - the isolation distance, which provides a quantitative measure of isolation quality.
Quality - a qualitative measure of isolation quality, which can be used to compare across different recording systems (isolation distances cannot be compared between e.g. a tetrode and an octrode).
Notes - user comments, e.g. about unresolved similarity between clusters.
This can be done by right clicking on the colored squares in the Cluster Panel, which brings a dialog where the information can be entered.
This information can be displayed in the Cluster Panel by selecting -> (selecting this menu item a second time will toggle its status and show the default layout).
This information is also shown in the status bar when the mouse is over a cluster in the Cluster Panel.
| Prev | Home | Next |
| Automatic Reclustering | Up | Saving the Cluster File |